The novel Coronavirus has been isolated countless times from patients, incl. “BavPat1/2020”, from one of the first patients in Bavaria, by the team around @c_drosten .
Several isolates can be ordered from bio material repositories, e.g.
The genomes of these isolated and laboratory grown viruses were sequenced, as were thousands of samples from patients around the globe: http://nextstrain.org/ncov/global.
By sequencing it is possible to clearly identify which exact virus is present.
By the way it’s still little known that diagnostic laboratories often analyze patient samples (i.e. swab material) also by sequencing in order to not solely rely on PCR
– especially if the PCR is ambiguous and/or the patient is in a bad condition!
Viruses are extremely small (10-100 times smaller even than E. coli bacteria!)- so how do you know if the virus will multiply? There are many different ways to find this out, which are also used for mutual confirmation. And now it gets exciting!
Even without fancy technology, such as PCR or electron microscopy, virus replication is completely unambiguous and can be recognized by everyone:
SARSCoV2 is very aggressive and kills infected cells rapidly during its reproduction! The more virus in the inoculum, the faster.
The pictures show lung cells (A549-ACE2) infected with SARS-CoV2, such that roughly every 10th cell was infected (first two pictures): after 1.5 days most cells are dead! If you use 10 times more virus, about half a day is already enough!

This extreme multiplication can also be found with other detection methods. By RT-PCR the amount of virus genomes in the cells can be measured / “counted”. In the beginning (2h) the added inoculum is measured, but already 2 hours later the number has increased a hundredfold!

Please note: the above diagram on the #PCR measurement has a logarithmic Y-axis, #RNA amplification (initially) is #exponential, i.e. EXTREMELY fast! You’ll find a very similar behavior when you measure the amount of viruses that are now produced and shed by these cells:
These newly formed viruses can of course be applied to lung cells again to confirm that they are “viable” viruses: these cells will also die! By diluting the virus and counting the “holes” in the cell layer, one can determine their number (see diagram above).
All three procedures show: #SARSCoV2 is extremely fast and aggressive, even compared to other viruses we work with. If it behaves even remotely similar in the lung (and it most likely does), you would rather not catch it!
Therefore and because we do not have good treatment options yet, we exclusively work in biocontainment labs of biosafety level 3 (4 would be the highest, e.g. for Ebola). This is no fun, even though
@VladimirMagalh looks quite cheerful in the photo!
Just: it makes a lot of sense!
Seeing is believing!
So can one also see the Coronavirus?
Yes, you can!
With much effort!
The viruses grown in the lab can now be visualized with an expensive electronmicroscope…
This is what’s happening here in #Heidelberg for example
in the group of Prof. #Bartenschlager
@UniHeidelberg
together with
@BriggsGroup
@embl
On the pictures below you can see the round virus envelope, inside the RNA genome (“knobs“), and on the outside the notorious spike protein by which the virus enters the cell.
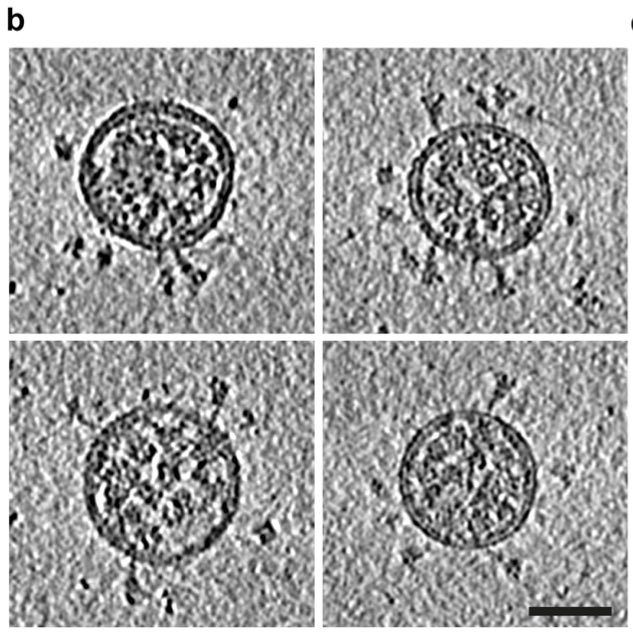
By capturing #virus particles from many different angles and with lots of computer power, you can even create detailed 3D views that provide details almost down to the single atom level! You can even look inside the virus and see…
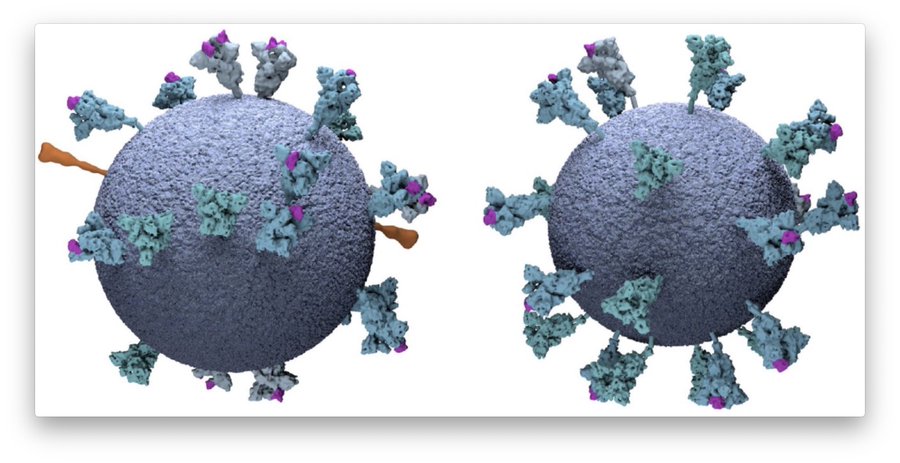
how efficiently the RNA genome, which is extremely long in relation to the virus particle, is packaged without the strand become knotted and thus unusable. This work was not done in Heidelberg but in the laboratory of
@drlisai
With this, I hope I could give some interesting „behind the scenes“ of experimental lab research, and a quick summary of “real” #virology on the new #Coronavirus #SARSCoV2.
Gladly RT and please note the sources in the comments!

